On May 25th, 2016, Nature, the top academic journal, online published the research progress of N6-adenosine methyltransferase complex, made by research team lead by professor Yin in National Key Laboratory of Crop Genetic Improvement /School of Biology Science and Technology. Headlined by Structural basis of N6-adenosine methylation by the methyltransferase-like 3(METTL3) and methyltransferase-like 14 (METTL14), this work reported the crystal structures of METTL3-METTL14 heterodimer, which provided the mechanism insight into RNA N6-methyladenosine modification, and served as an important foundation for understanding m6A epitranscriptomics.

As is known to all, RNA has central roles in biological system. Recently, chemical modifications of RNA has been the research focus. N6-methyladenosine is a prevalent RNA modification in species including viruses, bacteria, yeasts, plants and mammals. It functions in multiple aspects of developmental regulation by affecting aspects of RNA metabolism. In human, two MTases, METTL3 (also known as MT-A70) and METTL14 participate in this modification as the core components of the MTase complex.
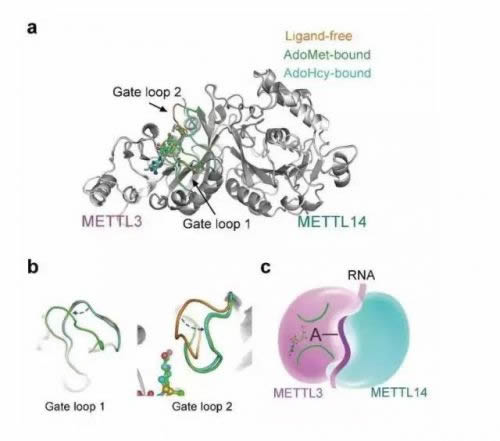
About 20 years ago, only a 70 kD protein was identified and named MT-A70 or METTL3, and the additional components, METTL14 were identified just two years ago. Why is this modification completed by two proteins and how do they function? Until recently, the molecular mechanism is still unknown. By determining the crystal structure of the core METTL3-METTL14 complex, this was elucidated.
Notably, S-adenosyl methionine (AdoMet) was observed in only the METTL3 pocket and not in METTL14, though both of them adopt a class I MTase fold. Combined with biochemical analysis, these results suggest that in the m6A MTase complex, METTL3 primarily functions as the catalytic core, while METTL14 serves as an RNA-binding platform. This structural information provides an important framework for the functional investigation of m6A.
This research was fulfilled by Ping Yin research team. Xiang Wang, Jing Feng and Yuan Xue are the joint first authors, and Ping Yin is the corresponding author. Our university platform, Central for Protein Research, provided strong technical support and the insect cell expression platform played a key role. Chun Tang, researcher of Nation Center for Magnetic Resonance, associated with this work. And all the data was collected in SSRF beamline BL17U, BL19U1, and BL19U2. This work was supported by funds from the Ministry of Science and Technology, Fok Ying-Tong Education foundation, the Fundamental Research Funds for the Central Universities, and Huazhong Agricultural University Scientific & Technological Self-innovation Foundation.