Working together with Guangxi University , Chen Lingling and Yang Qingyong from the research group of bioinformation team in Huazhong Agricultural University(HZAU) publish a paper under the title of "BnPIR: Brassica napus Pan-genome Information Resource for 1,689 accessions" in a botanical journal called Plant Biotechnology Journal, establishing the first bioinformation platform of Brassica napus (B. napus) pan-genome and comparative genomes.
According to the team leader, serving as the “standard map” of biological and scientific study, reference genome has promoted the significant production of animals and plants,the identification of variable sites related to economic characters and the discovery and application of key genes. However, under the influence of long-term evolution and artificial selection, various strains of the same species have accumulated abundant variation at the genetic and phenotypic levels, and a single reference genome often fails to contain abundant genetic variation information in the species. In other words, for most of species, many “ambiguous and unknown” areas exist in the “standard map” that we are using now. So we face lots of barriers in discovering and utilizing the genetic loci and genes related to important characters. In order to overcome these difficulties, scientists put forward the strategy to build pan-genome by integrating the resources of multiple representative species.
As a very “young” polyploid plant, B. Napus which contains abundant genetic variations was originally formed 7,500 years ago by the natural hybridization between Brassica rape and Brassica oleracea. In order to uncover these variations, based on the genome sequences of 8 B. napus species, the rapeseed team and bioinformation team of HZAU seek for collaboration and successfully analyze and identify many single nucleotide polymorphisms(SNP) and presence/absence variations(PAV) and build a 1.8GB pan-genome which contains nearly 150,000 genes. The researchers further combine a collection of 1688 rapeseed re-sequencing data and construct a comprehensive database, B. napus pan-genome information resource (BnPIR) for rapeseed researchers to quickly query and use relevant resources.
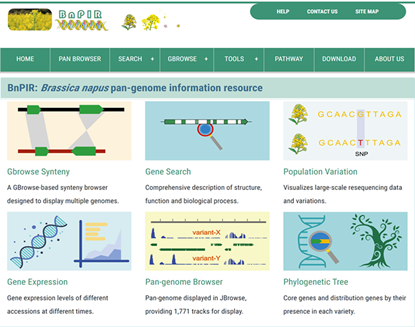
Based on gene information module, with Pan-genome Browser and Gbrowse Synteny as the core, BnPIR contains multi-omics data and common bioinformatics tools. BnPIR contains genomic sequences, gene annotations, phylogenetic relationship, expression data, PAV information, gene classification and common multi-omics tools and provides an integration of quick search and visualization. BnPIR will be a rich resource for rapeseed molecular biology and breeding, which will help rapeseed researchers to search and visualize their results in a pan-genome context, and provide a valuable template for pan-genome analyses in other species.
The research is completed through the cooperation between HZAU and Guangxi University. The pre-appointed Associate Professor Song Jiaming from Guangxi University and doctoral student Liu Dongxu from HZAU are co-first authors of the paper. Prof. Chen Lingling and Associate Professor Yang Qingyong are co-corresponding authors of the paper. The research has been funded by many projects, such as National Natural Science Foundation of China and Innovation Group of Natural Science Foundation of Hubei Province.
Paper link: https://doi.org/10.1111/pbi.13491
Source: http://news.hzau.edu.cn/2020/1020/58473.shtml
Translated by: Xiong Ying
Supervised by: Zhang Juan